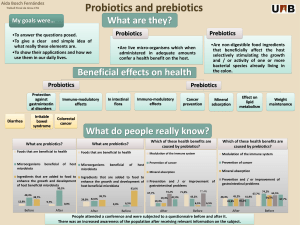
Fish Gut Microbiota: Composition, Factors & Health Impact
Telechargé par
Narimane Azouzi

See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/280005494
The Gut Microbiota of Fish
Chapter · October 2014
DOI: 10.1002/9781118897263.ch4
CITATIONS
19
READS
5,509
3 authors, including:
Some of the authors of this publication are also working on these related projects:
The Gut and Health Group at The Aquaculture Protein Centre (www.apc-coe.no) View project
Evaluation of cost-effective feed additives as immunostimulants in Aquaculture View project
Jaime Romero
University of Chile
159 PUBLICATIONS2,721 CITATIONS
SEE PROFILE
Einar Ringø
UiT The Arctic University of Norway
209 PUBLICATIONS9,453 CITATIONS
SEE PROFILE
All content following this page was uploaded by Einar Ringø on 15 October 2017.
The user has requested enhancement of the downloaded file.

TrimSize 170mm x 244mm Merrifield c04.tex V3 - 07/23/2014 10:31 A.M. Page 75
4 The Gut Microbiota of Fish
Jaime Romero1, Einar Ringø2and Daniel L. Merrifield3
1Instituto de Nutrición y Tecnología de los Alimentos (INTA), Universidad de Chile,
Santiago, Chile
2Norwegian College of Fishery Science, UiT The Arctic University of Norway,
Tromsø, Norway
3School of Biological Sciences, Plymouth University, UK
ABSTRACT
Animals harbour a complex microbial community, consisting of bacteria, yeast, viruses,
archaeans and protozoans, in their gastrointestinal (GI) tract. These microbes inuence
various host functions including development, digestion, nutrition, disease resistance and
immunity. One important aim of GI microbiota studies therefore is to give a scientic basis
for developing effective strategies for manipulating GI microbial communities to promote
the host health and improve productivity. This chapter reviews the current knowledge on
the microbiota composition in several sh species, emphasizing the compilation of results
reported regarding the most frequently observed bacterial genera and phyla in marine and
freshwater species. This also includes descriptions of the microbiota in early stages of
development, the inuence of environmental and host factors on the establishment of the
bacterial populations that become part of the gut microbiota, and the importance of these
microbial communities on host health, development and nutrition.
4.1 INTRODUCTION
In the classic description, the complex community of microorganisms inhabiting body sites in
which surfaces and cavities are open to the environment is termed the microbiota; previously
this was called the microora or microbial biota. Moreover, the epithelial surfaces of sh and
all other vertebrates are colonized at birth by large numbers of microorganisms (microbiota)
that form commensal or mutual relationships with their hosts (Spor et al. 2011). The majority
of these microbes reside in the digestive tract, where they inuence a broad range of host
biological processes. The vertebrate gut harbours a coevolved consortium of microbes that play
critical roles in the development and health of this organ. This microbial community can be
subcategorized into two major groups. One group simply passes through the lumen with food
Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics, First Edition. Edited by Daniel Merrield and Einar Ringø.
© 2014 John Wiley & Sons, Ltd. Published 2014 by John Wiley & Sons, Ltd.

TrimSize 170mm x 244mm Merrifield c04.tex V3 - 07/23/2014 10:31 A.M. Page 76
76 Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics
Fig. 4.1 Scanning electron micrograph of the anterior intestinal mucosa of rainbow trout; a pair of
autochthonous bacterial rods is present in close association with the mucosal brush border. Scale bar =
2μm. (Source: Merrifield et al. 2009. Reproduced with permission of John Wiley & Sons.)
or digesta (the allochthonous microbiota), whereas the other group is potentially resident and
intimately associated with host tissues (the autochthonous microbiota; Figure 4.1) (Ringø and
Birkbeck 1999). The normal microbiota has also been dened as the community of microbes
present in most individuals of a population or a species that, despite continual contact with
different tissues, cause no harm to the host (Berg 1996).
In previous investigations to study the microbiota of the GI tract of shes, the general
approach has been the use of conventional culture based methods (Cahill 1990; Ringø and
Birkbeck 1999). However, it has been reported that these methods present several disadvan-
tages since the number and species of bacteria detected are affected mainly by the culture
conditions and the culture media used, particularly certain fastidious and obligate anaerobes
(Spanggaard et al. 2000). These conventional methods are time consuming and lack accuracy
in isolate identication. Early in the 1990s, Cahill (1990) reviewed the current knowledge
concerning the bacterial communities in shes, at that time mostly based on culture-dependent
observations. The description provided in that review was mainly based on biochemical identi-
cation of the microorganisms, which has restricted discrimination power and may lack proper
denitions of relationships between aquatic-environmental microorganisms and sh micro-
biota. The lack of cultivability of the majority of the indigenous bacteria in many aquatic
environments, including the GI tract of aquatic animals, is becoming increasingly apparent
(Amann et al. 1995; as discussed in Chapter 5). For example, in Atlantic salmon (Salmo
salar L.), coho salmon (Oncorhynchus kisutch) or yellowtail (Seriola lalandi), cultivable bac-
teria (using tryptic soy agar (TSA) incubated at 10 days at 17 ∘C) represent ≤1% of the
total bacteria (Romero and Navarrete 2006; Navarrete et al. 2009; Aguilera et al. 2013). To

TrimSize 170mm x 244mm Merrifield c04.tex V3 - 07/23/2014 10:31 A.M. Page 77
The Gut Microbiota of Fish 77
study such environmental samples, several culture-independent molecular techniques have
been developed. These methods have allowed the identication of microorganisms without iso-
lation and the determination of the phylogenetic afliation of community members, revealing
the enormous extent of microbial diversity. The analysis of DNA extracted directly from a com-
plex environmental sample provides a powerful and relatively bias-free alternative approach
towards characterizing a microbial community (Nayak 2010). Typically, fragments of 16S or
18S ribosomal genes are selectively amplied by PCR to provide information on prokaryotic
and eukaryotic communities, respectively (Navarrete et al. 2010a). Patterns of diversity and
relative abundance of amplied DNA fragments can then be assayed using several strategies
(see Chapter 5). Massive sequencing technologies have revolutionized this eld by allowing
direct sequencing of millions of DNA molecules from a single sample (Qin et al. 2010). It is
thus now possible to obtain unbiased qualitative and quantitative reconstructions of complete
microbial communities – including both cultivable and uncultivable representatives – within
reasonable time frames and at affordable cost. Molecular techniques have been successfully
applied in dozens of studies proling the GI microbial communities of sh. Several attempts
have been made to describe the microbiota in a number of important aquacultured sh species.
Molecular methods based on PCR amplication of DNA extracted from frozen samples have
typically been the favoured approach and have proven to be efcient in studying the GI bac-
terial community of shes (Grifths et al. 2001; Jensen et al. 2004; Romero and Navarrete
2006; Hovda et al. 2007; Kim et al. 2007). Recently, studies have begun to analyse the sh
gut microbiota using massive sequencing strategies (e.g. van Kessel et al. 2011; Roeselers
et al. 2011; Desai et al. 2012; Wu et al. 2012). It is anticipated that as this technology becomes
more accessible it will signicantly improve our knowledge of the sh gut microbiota, enabling
identication of the rare biosphere and community metabolic pathways.
The ultimate goal of these studies is to provide a scientic basis for developing effective
strategies for manipulating gut microbial communities to promote animal health and improve
productivity. To achieve this goal, the principles governing microbiota composition (assembly)
and maintenance within the intestine must be understood. The vast majority of these studies
have focused on the bacterial communities and to a lesser extent yeast; very little information
is available for the viral, archaean and protozoan populations in the GI tract of sh.
4.1.1 Current knowledge of the gut microbiota in fish
Our current knowledge of the microbiota composition is derived from a compilation of
information in numerous reports; most of them focused on farmed sh, and among these the
salmonids have received much attention. Figure 4.2 summarizes the most commonly reported
bacterial phyla in salmonids, based on the review of Nayak (2010). Proteobacteria and
Firmicutes are the most frequently reported phyla in the salmonid gut microbiota, suggesting
that members of these bacterial classes are especially well adapted to conditions in the sh
intestine or their surrounding aquatic environment. Interestingly, studies in salmonids show
that some particular bacterial genera can be predominant in the microbiota composition; for
example, Pseudomonas can represent more than 60% of the community when ribosomal
amplicons are cloned and sequenced (Navarrete et al. 2009). The dominance of a particular
bacterial group has been observed in salmonid guts using similar culture-independent meth-
ods. Holben et al. (2002) reported that some genera were highly abundant in reared Atlantic
salmon from two different locations: in a Scottish hatchery, Mycoplasma corresponded to
81% of clones retrieved, whereas in a Norwegian hatchery, Acinetobacter accounted for 55%.

TrimSize 170mm x 244mm Merrifield c04.tex V3 - 07/23/2014 10:31 A.M. Page 78
78 Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics
0246
Number of reports
Phylum reported in salmonids
Proteobacteria
Firmicutes
Actinobacteria
Bacteroidetes
Fusobacteria
Tenericutes
Deinococcus-Thermus
810 12
Fig. 4.2 Bacterial phyla observed in the gut microbiota of salmonids. (Source: Nayak 2010.
Reproduced with permission of John Wiley & Sons.) For colour detail see Plate 9.
Although other genera were also present, their abundance was closer to 2%. Interestingly, in
wild salmon (entirely carnivorous), the abundance of Mycoplasma was 96% of the clones
analysed. Similarly, Pond et al. (2006) described the intestinal microbiota of rainbow trout
(Oncorhynchus mykiss Walbaum) by using a cloning approach. They reported only two major
groups among 200 clones analysed, which corresponded to Clostridium and Aeromonas.
Furthermore, Kim et al. (2007) reported that Clostridium dominated the gut microbiota in
rainbow trout analysed by denaturing gradient gel electrophoresis (DGGE). The carnivorous
diet of salmon may explain in part the low number of taxa observed, since a recent study
indicated that diet inuences the bacterial diversity of the digestive tract. In this report,
a more comprehensive analysis of vertebrate gut microbiota (albeit mostly mammalian)
indicates that bacterial diversity increases from carnivore to omnivore to herbivore (Ley
et al. 2008). This has recently been observed in Antarctic sh, among which the omnivorous
Notothenia coriiceps (yellowbelly rockcod or bullhead notothen) exhibits greater diversity
than the exclusively carnivorous Chaenocephalus aceratus (blackn or Scotia Arc icesh)
(Ward et al. 2009). This may indicate that increasing herbivory in sh leads to gut microbiota
diversication, as observed in mammals.
Descriptions of microbiota from wild sh have also been reported, especially in herbivorous
sh and also in some habitats of ecological interest. Recently, the microbiota of three sh from
a coral reef was reported (Smriga et al. 2010). The studied sh corresponded to different diets
representing two trophic levels: Chlorurus sordidus (parrotsh) is a herbivore that consumes
primarily endolithic and epilithic algae; Lutjanus bohar (two-spot red snapper) is a top preda-
tor that consumes shes and crustaceans; and Acanthurus nigricans (whitecheek surgeonsh)
 6
6
 7
7
 8
8
 9
9
 10
10
 11
11
 12
12
 13
13
 14
14
 15
15
 16
16
 17
17
 18
18
 19
19
 20
20
 21
21
 22
22
 23
23
 24
24
 25
25
 26
26
 27
27
1
/
27
100%