ucalgary_2014_elkadiri_zineb.pdf

UNIVERSITY OF CALGARY
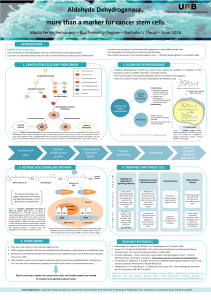
Epoxyeicosanoids (EETs) Promote The Cancer Stem Cell State by Preventing Breast Cancer
Cells Differentiation
by
Zineb El Kadiri
A THESIS
SUBMITTED TO THE FACULTY OF GRADUATE STUDIES
IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE
DEGREE OF MASTER OF SCIENCE
DEPARTMENT OF BIOLOGICAL SCIENCE
CALGARY, ALBERTA
MAY, 2014
© Zineb El Kadiri 2014

Abstract
Our goal is to examine the role of epoxyeicosatrienoic acids (EETs) – omega-6 poly-unsaturated
fatty acid metabolites - in modulating breast cancer cell plasticity manifest in the discrete
attractor state transitions between cancerous and differentiated states. We used an in vitro breast
cancer cell differentiation system, MDA-MB231 that exhibited two quasi-discrete
subpopulations representing a stem-like and a differentiated state. We found that elevated EETs
suppressed the vitaminD-induced shift of the MDA-MB231 cells towards the differentiated state.
Moreover, subpopulation gene expression profiles of cells exposed to EETs robustly identified
stem cell-like and differentiated subpopulations as two discrete clusters, regardless of EETs.
Interestingly, EETs promoted “stemness” and metastasis genes, while suppressed differentiation
related genes, suggesting that EETs not only blocked the switch between the two attractors but
also affected cell-intrinsic attractor properties. Our findings suggest that anti-EETs agents may
have therapeutic or preventive utility against breast cancer that could be further explored.
ii

Acknowledgements
I wish to thank my thesis supervisor, Dr. Sui Huang for the opportunity to participate in
this research project. I am grateful for his mentoring and patience as we explored new ideas
together and defined the direction of my research. I would like to thank all those who have
contributed to my work and supported me during my study including, Wei Wu, Mitra Mojtahedi,
Ivan Gomez and Rebecca Leong. A special thank to Dr. Joseph Zhou for his help in analyzing
the microarray data.
The work in this thesis would not have been possible without the generous help and
support I have received from my husband, my little ones Ilyas and Sulayman, my mom and
sisters. Thank you all for your unwavering love and encouragement.
iii

Table of Contents
Abstract............................................................................................................................... ii
Acknowledgements............................................................................................................ iii
Table of Contents............................................................................................................... iv
List of Tables .................................................................................................................... vii
List of Figures.................................................................................................................. viii
List of Symbols, Abbreviations and Nomenclature........................................................... xi
CHAPTER ONE: INTRODUCTION..................................................................................1
1.1 Systems view of the gene regulatory network: cancer cells as cancer attractors ......1
1.1.1 System biology approach ..................................................................................1
1.1.2 Theory of gene regulatory network and attractors ............................................2
1.1.3 Abnormal cell types as cancer attractors ...........................................................4
1.1.4 The concept of cancer stem cell: refining of cancer cell hypotheses vs. clonal
evolution ............................................................................................................6
1.2 Comparing the normal stem cells (NSC) with the cancer stem cells (CSC): the
hallmark properties, the cell of origin and the surrounding microenvironment ......7
1.2.1 Normal stem cells: self-renewal and differentiation potential ..........................8
1.2.2 The cancer stem cell of origin ...........................................................................9
1.3 Breast cancer is a stem cell disease that could be influenced by environmental factors
..................................................................................................................................9
1.3.1 Environmental factors account to a certain degree for breast cancer disparities9
1.3.2 Mammary stem cells in normal development .................................................11
1.3.3 Mammary cancer stem cells in solid tumors ...................................................12
1.3.4 Breast cancer is a disease with functional inter-tumor and intra-tumor
heterogeneity....................................................................................................13
1.3.4.1 Breast cancer cell population heterogeneity is reflected by their stem cell
marker expression: ..................................................................................15
1.3.4.2 Functional heterogeneity in CD44 high human breast cancer stem cell like
compartment: ..........................................................................................16
1.3.4.3 CD44+/CD24-pattern correlates with the basal breast cancer subtype rather
than tumorigenisis:..................................................................................17
1.4 Molecular biology of breast cancer .........................................................................19
1.4.1 Cell signalling pathways regulating self-renewal and differentiation of Breast
normal and cancer stem cells ...........................................................................19
1.4.1.1 The canonical Wnt signalling pathway:.................................................20
1.4.1.2 The Notch signalling pathway: ..............................................................21
1.4.1.3 The Hedgehog signalling pathway: .......................................................22
1.4.1.4 The PTEN PI3K/Akt signalling pathway: .............................................22
1.4.2 Cancer stem cell resist therapeutic challenges and drive the relapse of the initial
heterogeneous tumor........................................................................................23
1.4.3 Basal subclass of breast cancer is associated with high recurrence rate and
metastatic spread..............................................................................................24
iv

1.5 Induced differentiation of human breast cancer cells reflects the dynamics of non-
genetic heterogeneity .............................................................................................26
1.5.1 Disrupting the immature state: differentiation of breast cancer cells..............26
1.5.2 Vitamin D metabolism and mechanism of action ...........................................28
1.5.3 The role of vitamin D in repressing the progression and promoting differentiation
of basal subtype of breast cancer cells.............................................................29
1.6 The cytochrome P450-derived eicosanoids (EETs) pathway in conjunction with cancer
................................................................................................................................30
1.6.1 Epoxyeicosatrienoic acids: formation, metabolism, and mechanism..............31
1.6.1.1 Production of EETs:...............................................................................32
1.6.1.2 Mechanism of action of EETs: ..............................................................34
1.6.1.3 Metabolism of EETs: .............................................................................35
1.6.2 EETs signalling in cancer................................................................................35
1.6.2.1 High levels of EET induce the activation of pathways involved in
proliferation, angiogenesis and anti-apoptosis:.......................................35
1.6.2.2 Synthetic EETs trigger tumor initiation and metastasis in animal models:
.................................................................................................................38
CHAPTER TWO: GOALS AND APPROACHES OF THE PROJECT ..........................40
2.1 Overall goal and biological rationale.......................................................................40
2.2 Experimental system................................................................................................44
2.2.1 MDA-MB-231 cell line as a surrogate for primary breast tumor cells ...........44
2.2.2 Vitamin D-induced differentiation highlights the micro-heterogeneity in the
clonal population of MDA-MB-231 cell line ..................................................45
2.2.3 Decreased aldehyde dehydrogenase functional activity as a marker for breast
cancer cells differentiation...............................................................................46
2.2.4 Direct and indirect pharmacological manipulation of EET levels ..................48
CHAPTER THREE: MATERIALS AND METHODS ....................................................53
3.1 Cell culture...............................................................................................................53
3.2 Preparing vitamin D and soluble epoxide hydrolase inhibitor stock solutions........53
3.3 Vitamin D induced differentiation system...............................................................53
3.4 MTT proliferation assay ..........................................................................................54
3.5 Western blot analysis ...............................................................................................55
3.6 Flow cytometry analysis ..........................................................................................55
3.6.1 Aldehyde dehydrogenase activity distribution using flow cytometry.............55
3.6.2 7-amino actinomycin D staining for detecting apoptotic cells........................56
3.7 Statistical analysis:...................................................................................................56
3.8 Microarray analysis..................................................................................................57
3.8.1 FACS sorting and RNA isolation:...................................................................57
3.8.2 Microarray analysis .........................................................................................57
3.9 Methodology: Gene Expression Dynamics Inspector (GEDI) ................................58
3.9.1 Introduction of GEDI ......................................................................................58
3.9.2 Philosophy of GEDI ........................................................................................59
3.9.3 Data presentation of GEDI ..............................................................................60
3.9.4 Self-organizing map and GEDI .......................................................................60
3.9.5 Gene Sets Enrichment Analysis (GSEA) ........................................................61
v
 6
6
 7
7
 8
8
 9
9
 10
10
 11
11
 12
12
 13
13
 14
14
 15
15
 16
16
 17
17
 18
18
 19
19
 20
20
 21
21
 22
22
 23
23
 24
24
 25
25
 26
26
 27
27
 28
28
 29
29
 30
30
 31
31
 32
32
 33
33
 34
34
 35
35
 36
36
 37
37
 38
38
 39
39
 40
40
 41
41
 42
42
 43
43
 44
44
 45
45
 46
46
 47
47
 48
48
 49
49
 50
50
 51
51
 52
52
 53
53
 54
54
 55
55
 56
56
 57
57
 58
58
 59
59
 60
60
 61
61
 62
62
 63
63
 64
64
 65
65
 66
66
 67
67
 68
68
 69
69
 70
70
 71
71
 72
72
 73
73
 74
74
 75
75
 76
76
 77
77
 78
78
 79
79
 80
80
 81
81
 82
82
 83
83
 84
84
 85
85
 86
86
 87
87
 88
88
 89
89
 90
90
 91
91
 92
92
 93
93
 94
94
 95
95
 96
96
 97
97
 98
98
 99
99
 100
100
 101
101
 102
102
 103
103
 104
104
 105
105
 106
106
 107
107
 108
108
 109
109
 110
110
 111
111
 112
112
 113
113
 114
114
 115
115
 116
116
 117
117
 118
118
 119
119
 120
120
 121
121
 122
122
 123
123
 124
124
 125
125
 126
126
 127
127
 128
128
 129
129
 130
130
 131
131
 132
132
 133
133
 134
134
 135
135
1
/
135
100%