Sunflower Genetic Diversity in Tunisia: A Conference Paper
Telechargé par
agroecobio saad

International Workshop
Advances in Plant Biotechnology for Crop Improvement
INAT, 18-20 April 2016
Assessment of genetic diversity for subsequent improvement of confection sunflower in Tunisia
Khoufi Sahari1*, Ben Jeddi Fayçal1,Brunel Dominique2
1 Laboratoire des Sciences Horticoles, Institut National Agronomique de Tunisie, 43, avenue Charles Nicolle 1082 Tunis-Mahrajène Tunisie. *: sk111[email protected]
2 US Etude du Polymorphisme des Génomes Végétaux, INRA, CEA/IG/Centre National de Génotypage, Université Paris Saclay, 2 rue Gaston Crémieux, 91057 Evry Cedex, France.
Introduction
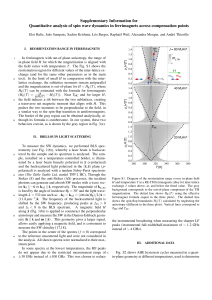
Sunflower (Helianthus annuus L.) is cultivated and consumed in Tunisia mainly as confectionery seeds, which correspond to cultivars with
particular phenotypic characteristics (seed size, 100 seed weight, color…). We focused on studying the genetic variation among a collection of 59
populations of cultivated sunflower in Tunisia, combining SSR and SNP as molecular markers in order to start a new plant breeding program.
Plant material:
59 sunflower
populations (Fig. 1)
and 7 reference
varieties * (SF012,
SF193, SF085,
SF332, SF092,
SF109 Var Turk).
DNA extraction:
Total genomic
DNA was
extracted using
DNeasy 96
Plant Kit
(QIAGEN,
Valencia, CA).
SSR assay:
56 Primers
*covering 17
linkage groups.
SNP assay:
EFE,
EXECUTER1,
AVP1, CG068,
P5CS2, LPT3a
and LPT3b
genes*.
DNA sequencing:
was conducted
with Illumina
Sequencing
Systems.
Bioinformatic
analysis:
were
conducted with
CLC Genomics
Workbench
(v7).
Statisticle
analysis:
Factorial
Correspondence
Analysis (FCA)
was conducted
using XLSTAT
software (V1.06).
Core
collections:
was performed
with MSTRAT
software
version v4
(Gouesnard et
al., 2001).
Materials and methods
*: reference varieties, SSR markers and genes are
provided by the laboratory of Plant-Microbe
Interactions (LIPM) INRA Toulouse.
Results and Discussion
A total of 194 alleles were observed for thirty
microsatellite loci (3-10 alleles per locus) and 54
haplotypes corresponding to 117 SNPs identified
by NGS sequencing of seven candidate genes (4-
16 haplotypes per gene).
No relation was found between genetic and
geographical distance within Tunisian
populations (r = 0.08; p = 0.18), which suggests
seed exchanges between farmers, or agricultural
institutes.
MSTRAT generated a core collection: [Hat28 (Slougia), Hat15 (Dougga), Hat40 (Oued Zarga),
Hat20 (Ksar Mezouar), Hat25 (Ain Chelou), Hat43 (Sejane), Hat11 (Tounga), and Hat65 (Oued
Beja), capturing 90 % of molecular allelic content of the 59 populations (Fig. 3).
Fig. 3: Comparison of the effectiveness in sampling genetic
diversity between the M strategy (top curve) and the random
strategy (bottom curve), according to the number of populations
of the core collection. Molecular data (SSR and SNP) are the
“active variables.
Fig. 2: Distribution of reference varieties and Tunisian populations of
sunflower according to factors 1 and 2 of FCA based on SSR (a) and
SNP (b) markers.
Conclusion
This study highlighted the genetic originality of the Tunisian material compared to reference
varieties. No geographical structure was noted, however 8 populations were sorted out by SNP and
SSR markers representing a core collection for future improvement of sunflower.
References
Gouesnard, B., Bataillon, T.M., Decoux, G., Rozale, C., Schoen, D.J. and David, J.L., 2001. MSTRAT: An algorithm for building germplasm core collections by maximizing allelic or phenotypic richness. Journal of heredity 92: 93-94.
Acknowledgements: We thank Nicolas Pouilly, Aurélie Bérard for laboratory assistance,
Patrick,Vincourt, Stéphane Muños for data analysis and Brigitte Gouesnard for MSTRAT analysis
and discussions. Funding for this study was provided by the laboratory of Plant-Microbe
Interactions (LIPM) and US_EPGV (Etude du Polymorphisme des Génomes Végétaux) of the
French National Institute for Agricultural Research (INRA).
Factorial correspondence
analysis (FCA) highlighted
some genetic originality of
the Tunisian material
compared to six reference
varieties from different
countries (Fig. 2).
Fig. 1: Sites of sampled populations of sunflower (Helianthus annuus L.)
used in this study, circled population represent the core collection.
1
/
1
100%