Apports de la spectromètrie de masse en pathologie

Apports de la
spectromètrie de masse
en pathologie infectieuse
12es JNI TOULOUSE 8-10 JUIN 2011
Michel Drancourt, MD, PhD
URMITE UMR CNRS 6236 IRD 198 IFR 48
Institut Hospitalier et Universitaire POLMIT
Université de la Méditerranée, Marseille

CHU la Timone
Laboratoire de Bactériologie et de
Virologie
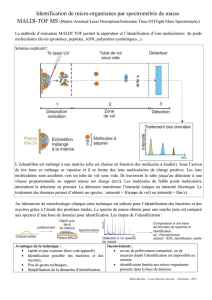
Structure générale d’un spectromètre de masse
SOURCE ANALYSEUR DETECTEUR
Pour volatiliser et
ioniser les molécules
Il mesure les valeurs
du rapport: masse / nb
de charge (appelé m/z)
Détection des
ions
+ -
détecteur

CHU la Timone
Laboratoire de Bactériologie et de
Virologie
Principe de la source MALDI
•Etapes d’ionisation
–1- Co-crystallisation de l’échantillon et de la matrice
photosensible
–2- Excitation des molécules de matrices par
l’impulsion laser
–3- explosion de la matrice et ionisation de la matrice
–4- transfert de charge et ionisation de l’échantillon
H+
Transfert de
charge
H+
Ionisation
MM
Analyte Analyte

CHU la Timone
Laboratoire de Bactériologie et de
Virologie
Le détecteur
ions
Amplification électronique
Signal
électrique
Principe d’amplification
1- un ion frappe le détecteur et arrache des électrons
2- les électrons arrachés frappent le détecteur et arrachent d’autres électrons, et ainsi de suite
3- l’ensemble des électrons arrachés vont forme un courant électrique

Pick-up
colony
Deposit colony on plate
MALDI-TOF processing
Data collection,
interpretation and reporting
MALDI-TOF plate loading /
matrix overlaying
Identification
Profil
Database
L’identification des colonies bactériennes et levures
 6
6
 7
7
 8
8
 9
9
 10
10
 11
11
 12
12
 13
13
 14
14
 15
15
 16
16
 17
17
 18
18
 19
19
 20
20
 21
21
 22
22
 23
23
 24
24
 25
25
 26
26
 27
27
 28
28
 29
29
 30
30
 31
31
 32
32
 33
33
 34
34
 35
35
 36
36
 37
37
 38
38
 39
39
 40
40
 41
41
 42
42
1
/
42
100%