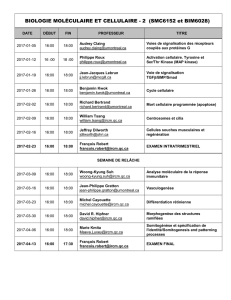
Meeting agenda - Institut de recherches cliniques de Montréal

Programme / Agenda
(Auditorium Jacques-Genest et salle André-Barbeau)
Mardi 2 avril - Tuesday, April 2
13 h Accueil des participants et installation des affiches - Attendee greeting and poster installation
13 h 45 Mot d’ouverture / Opening remarks
Tarik Möröy (Président et directeur scientifique /President and Scientific Director, IRCM)
Session 1: Biologie des Réseaux / Network Biology
(Président / Chair: Marlene Oeffinger, IRCM)
14 h John Aitchison, Institute for Systems Biology, Seattle Biomedical Research Institute
Nuclear Pore Complex Mediated Control of Chromatin
14 h 40 Thomas Kislinger, MaRS Centre, University of Toronto
Plasma Membrane Proteomics: Stem Cell Signatures & Insights into Cardiac Biology
15 h 20 Guillaume Diss, Université Laval (Superviseur / Supervisor: Christian R. Landry)
A Systematic Approach for the Genetic Dissection of Protein Complexes in Living
Cells
(Conférencier sélectionné parmi les résumés / Speaker selected from abstracts)
15 h 40 Pause-café et visionnement des affiches / Coffee break and poster viewing
(Hall d’entrée / Lobby)
16 h Michael Tyers, IRIC, Université de Montréal
Chemical Biology of the Ubiquitin System
16 h 40 Amy Caudy, Donnelly Centre, University of Toronto
Discovery in the Metabolic Network by Genomics and Full Scan Metabolomics
17 h 20 Benoit Coulombe, IRCM, Université de Montréal
Mapping the Human Methyltransferasome Reveals the Existence of a PTM Code that
Targets Molecular Chaperones to Regulate Functional Organisation of the Proteome
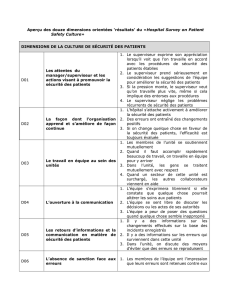
Mercredi 3 avril - Wednesday, April 3
Session 2: Génomique Médicale et Évolutive / Medical and
Evolutionary Genomics
(Président / Chair: Jacques Archambault, IRCM)
9 h Igor Jurisica, Ontario Cancer Institute, University of Toronto
Network-Based Characterization of Drug-Regulated Genes, Drug Targets, & Drug
Toxicity
9 h 40 Mike Hallett, The Rosalind and Morris Goodman Cancer Research Centre, McGill University
Prognosis in Breast Cancer: the Illusion of Accuracy
10 h 20 Pause-café /Coffee Break
10 h 40 Philip Awadalla, CHU Sainte-Justine Research Center, Université de Montréal
CARTaGENE: A Population Based approach to systems genomics of quantitative
traits
11 h 20 Nada Jabado, McGill University Health Center
When Genetics meet Epigenetics: A New Option for Therapy in Pediatric High-Grade
Astrocytomas?
12 h Benjamin Haibe-Kains, IRCM, Université de Montréal
Prognostic and Predictive Value of Random Biomarkers in Cancer
12 h 30 Lunch (salle 255)
13 h Session d’affiches / Poster Session
1

Programme / Agenda
(Auditorium Jacques-Genest et salle André-Barbeau)
Session 3: Biologie des ARN / RNA Biology
(Président / Chair: Eric Lécuyer, IRCM)
14 h Nahum Sonenberg, The Rosalind and Morris Goodman Cancer Research Centre, McGill
University
Translational Control of Cancer and Autism
14 h 40 Timothy Hughes, Donnelly Centre, University of Toronto
A Compendium of RNA-Binding Protein Motifs for Analysis of Gene Regulation
15 h 20 Daniel Zenklusen, Université de Montréal
Characterization of Antisense RNA-Mediated Transcriptional Gene Silencing in Yeast
(Conférencier sélectionné parmi les résumés / Speaker selected from abstracts)
15 h 40 Pause-café / Coffee Break
16 h Christopher Burge, Massachusetts Institute of Technology
Dynamics of Alternative Splicing and Translation
16 h 40 Jérôme Waldispühl, McGill Centre for Bioinformatics, McGill University
Exploring the RNA Mutational Landscape
17 h 20 Mot de conclusion et départ / Concluding remarks and departure
Francois Robert (Directeur, axe de recherche en biologie des sytèmes et chimie médicinale
/ Director, Systems Biology and Medicinal Chemistry research division, IRCM)
2

Séance d’affiches / Poster Session
(1) AGUILAR, Lisbeth C., IRCM (Superviseure:Marlene Oeffinger)
Determining the temporal order of pre-ribosomal assembly
(2) AYASHI, Sammy, Université de Montréal (Superviseur: Dindial Ramotar)
Identification of Histone variants defective in cellular response to the immunosuppressant and
anticancer drug rapamycin
(3) AZIZI, Afnan, Université d’Ottawa (Superviseur: Mads Kearn)
Titre à déterminer / Title to be determined
(4) BERGALET, Julie, IRCM (Superviseur: Éric Lécuyer)
Defining the role and the mechanism of RNA localization to the mitotic apparatus
(5) BERLIVET, Soizik, IRCM (Superviseure: Marie Kmita)
Role of chromatin organization in HoxA gene regulation
(6) CELTON, Magalie, IRIC, Université de Montréal (Superviseur: Brian Wilhelm)
Expression of GATA2 in normal karyotype acute myeloid leukemia
(7) CLÉROUX, Katherine, IRCM (Superviseur: Marlene Oeffinger)
Linking Regulation and Spatial Progression of Pre-Ribosomes
(8) CODY, Neal, IRCM (Superviseur: Éric Lécuyer)
Defining the role for mRNA localization in the regulation of epithelial cell polarity
(9) DOSTIE, Starr, IRCM (Superviseur: Yvan Guindon)
Diastereoselective Synthesis of Nucleoside Analogues via Synthesis and Cyclization of Acyclic
Precursors
(10) EAR, Po Hien, Lady Davis Institute (Superviseur: Stéphane Richard)
Sam68 regulates the alternative splicing of a key Notch signaling factor required for lung cancer
(11) HELNESS, Anne, IRCM (Superviseur: François Robert)
Effect of Gfi1 absence on the epigenome of mouse hematopoietic stem cells and
progenitor cells
(12) IAMPIETRO, Carole, IRCM (Superviseur: Éric Lécuyer)
mRNA nuclear Retention as a Novel Facet of the DNA Damage Response in Drosophila
(13) JERONIMO, Célia, IRCM (Superviseur: François Robert)
FACT and Spt6 Act as a Histone Triage Agents Preventing Accumulation of H2A.Z within
Transcribed Regions
(14) KODIHA, Mohamed & MAHBOUDI, Hicham, Université McGill (Superviseur: Ursula Stochaj)
Methods developed for the automated quantification of granular cell compartments reveal novel
biological properties of stress granules
(15) LEBEDEV, Anton, IRCM (Superviseur: François Robert)
Mapping and Quantifying the Transcriptome of Murine HSCs and Hematopoietic Progenitors
by RNA-Seq: The Role of the Transcription factor Gfi1
3

(16) LEE, Sangkyu, Université McGill (Superviseur: Issam El Naqa)
Assessment of different machine learning techniques for multivariate radiation pneumonitis
modeling
(17) LEGENDRE, Félix, IRCM (Superviseur: Éric Lécuyer)
Defining a novel mechanism of mRNA localization to centrosomes in Drosophila
(18) MOUJABER, Ossama, Université McGill (Superviseur: Ursula Stochaj)
Biomarkers for the Aging of Human Mesenchymal Stem Cells
(19) PHENIX, Hilary, Université d’Ottawa (Superviseur: Mads Kaern)
Identifability and Interence of Pathway Topology by Epistasis Analysis
(20) ROCHETTE, Samuel, Université Laval (Superviseur: Christian R. Landry)
“Pin” pointing the dynamic of protein interaction networks (PINs)
(21) SALEH, Sadiq M., Université McGill (Superviseurs: Morag Park & Michael Hallett)
Mouse Models Of Breast Cancer Identify Oncogene-Specific Stroma Associated With Breast
Cancer Molecular Subtypes
(22) TAMBUTET, Guillaume, IRCM (Superviseur: Yvan Guindon)
Second Generation of Nucleoside Analogues Targeting Solid Tumor
(23) ZHANG, Olivia, IRCM (Superviseurs: Éric Lécuyer & Jérôme Waldispühl)
Define structured, unstructures and disordered regions in coding RNA sequences
(24) ZHOU, Gina, Université McGill (Superviseur : David Juncker)
Cost-effective multiplexed immunoassays with fepto-molar sensitivity using silver
amplification and a desktop scanner
4
1
/
4
100%