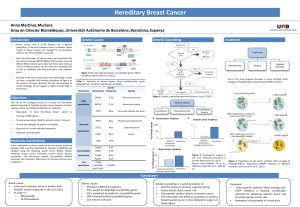
R links Download and Install latest R version click on

R links
Download and Install latest R version
http://cran.r-project.org/bin/macosx/
click on R-3.2.0.pkg, save the file and open it, simply follow installation process.
R is automatically installed on your Application folder
Quick-R (relatively) user-friendly guide to R
http://www.statmethods.net/
The StackOverflow forum is a trustworthy source to solve problems with R
(just google “R <your problem>” most of the times entries from StackOverflow come first)
---------------------------------------------------------------------------------------------------------------------
R package to retrieve and access data from the cBioPortal
http://www.cbioportal.org/cgds_r.jsp
To install from R:
Packages & Data → Package Installer → cgdsr
(click Install Dependencies)
# import package in R
library(cgdsr)
Useful functions:
● CGDS creates a connection with the server
● getCancerStudies returns a table of available cancer studies
● getCaseLists given a cancer study, returns available case lists
● getGeneticProfiles given a cancer study, returns available genetic profiles
● getProfileData retrieve the data for a specific study, case list, and genetic profile
See the documentation for a detailed description of these function.

Example:
# create connection to the server, you will always need to specify this connection in input
mycgds = CGDS("http://www.cbioportal.org/public-portal/")
# retrive cancer studies
studies = getCancerStudies(mycgds)
# print the table (without row numbers and quotes)
write.table(studies,file="cancer_studies.txt",sep="\t",row.names=F,quo
te=F)
# select a cancer study by ID (Column 1 in studies gives you all the IDs)
mycancerstudy = "prad_tcga"
# retrieve all case lists for a given study
allLists = getCaseLists(mycgds, mycancerstudy)
# select a list of cases for your cancer study (getCaseList returns you a table with IDs and
description of all case lists, you’re selecting here the first element of the first column)
mycaselist = getCaseLists(mycgds, mycancerstudy)[1,1]
# tables/matrices are accessed by row and column indices in square brackets
# matrix[2,1] element in the second row and first column
# matrix[1,] all elements in the first row
# matrix[,4] all elements in the fourth column
# matrix[1:3,4:6] elements between row 1 to 3 and columns 4 to 6
# matrix[c(1,3),c(4,6)] elements in row 1 and 3 and columns 4 and 6
# retrieve all genetic profiles for a given study
allProfiles = getGeneticProfiles(mycgds, mycancerstudy)
# select a genetic profile (getGeneticProfiles returns you a table with all genetic profiles
available for your selected study)
mygeneticprofile = "prad_tcga_gistic"
# retrieve the selected genetic profile for input genes
dataCNA = getProfileData(mycgds, c('BRCA1','MYC'), mygeneticprofile,
mycaselist)
# a vector is specified in R by the command c() with elements separated by commas
# vector = c(a,b,c)
# vector[2] = b

# select a second genetic profile
mygeneticprofile = "prad_tcga_rna_seq_v2_mrna"
# retrieve the selected genetic profile for input genes
dataRNA = getProfileData(mycgds, c('BRCA1','MYC'), mygeneticprofile,
mycaselist)
# merge the genetic data (and adjust row names of the table)
# you have now a table where each sample is a row and each column a gene specific data
# type (e.g. column 1 is BRCA1 copy number status)
data = merge(dataCNA, dataRNA, by="row.names")
rownames(data) = data$Row.names
data = data[,-1]
# plot the expression of a gene with respect to its copy number status
boxplot(data$BRCA.y ~ data$BRCA.x)
boxplot(data$MYC.y ~ data$MYC.x)
---------------------------------------------------------------------------------------------------------------------
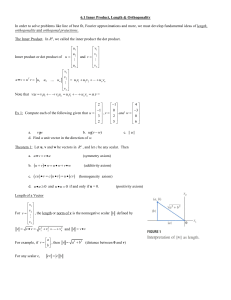
# Load tabular data using read.delim / read.table, my table has a header
data = read.delim(“my_table.txt”, header = T)
# Here I specify that the first column should be interpreted as row names (row.names = 1)
# vice versa the first row is NOT a header (header = F)
# if my table doesn’t have a header, I can define a header myself by giving column names
# col.names = c(...)
data = read.table(“my_table.txt”, row.names = 1, header = F,
col.names = c(“col_A”,“col_B”));
# Here my table has both a header and row names
# however I want to prevent R from modifying column names,
# e.g. by converting ‘-’ into ‘.’ (default behaviour in R)
# to do that: check.names = F
data = read.table(“my_table.txt”, row.names = 1, header = T,
check.names = F);
# subsetting by list of values
sub.data = data[data$cell.name == “A” | data$cell.name == “B” |
data$cell.name == “C”,]

# alternatively
selected = c(“A”,“B”,“C”)
sub.data = data[data$cell.name %in% selected,]
# if I want to extract the tissue types of the selected cell lines
tissues = data$tissue[data$cell.name %in% selected]
# NOTE I am not adding the ‘,’ before the last parenthesis → the comma indicate the extra
# dimension which in this cases there is not (data is a table, data$tissue is a vector)
# plot data
# the plot function to compare to continuous variables
plot(data$BRCA1, data$BRCA2)
# explore plot parameters
plot(data$BRCA1, data$BRCA2, pch=19, col=”red”, cex=0.5)
# boxplot to compare continuous variables with categorical ones (i.e. expression versus copy
# number category)
boxplot(data$BRCA1_exp ~ data$BRCA1_cna)
1
/
4
100%


![Poster LIBER san antonio 2011 [Mode de compatibilité]](http://s1.studylibfr.com/store/data/000441925_1-0f624c1012097e18f69fca01a2951eb6-300x300.png)