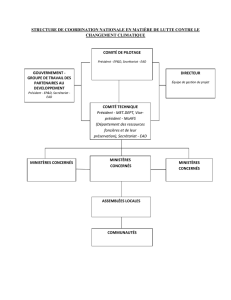
appel de la fonction principale et formatage de l`apres reponse

// appel de la fonction principale et formatage de l'apres reponse
function validate_form() {
txt = document.form.search.value;
jsi = new makeLinks(linksize);
searchLinks(jsi, txt);
}
//var key = "";
function makeEntry (){
this.Name="";
this.URL = "";
this.Desc = "";
this.Category = "";
this.Target = "";
return this;
}
function makeArray(n) {
this.length = n;
for (var k = 1; k <= n; k++) {
this[k] = "";
}
return this;
}
// Creation du tableau de la base
function makeLinks(size) {
this.length = size;
for (var r=1; r<= size; r++) {
this[r] = new makeEntry();
this[r].Name = namesArray[r];
this[r].URL = urlsArray[r];
this[r].Desc = descArray[r];
this[r].Category = categoryArray[r];
this[r].Target = targetArray[r];
}
return this;
}
// Description et indexation du tableau de la base
var linksize=0
datesArray = new makeArray(linksize);
namesArray = new makeArray(linksize);
urlsArray = new makeArray(linksize);
descArray = new makeArray(linksize);
categoryArray = new makeArray(linksize);
targetArray = new makeArray(linksize);
// Initialisation de la base

var arraycount=0
// Debut des donnees
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/index.html";
namesArray[arraycount] = "Cours de Biochimie et d'Enzymologie niveaux L1 - L2 -
M3";
descArray[arraycount] = "cours course lesson biochimie biochemistry structure
metabolisme metabolism glycolyse enzymologie enzymology chromatographie
chromatography radioactivite radioactivity radioprotection";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/1VELATP.h
tm/111VELATP.htm";
namesArray[arraycount] = "Energie libre de Gibbs - ATP";
descArray[arraycount] = "variation energy free atp adenosine adenosin triphosphate
phosphate constante constant equilibre equilibrium thermodynamique
thermodynamic bioenergetique premier principe principle interne enthalpie enthalpy
spontane spontaneite processus reversibilite reversibility reversible second entropie
entropy etat state standard";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/1VELATP.h
tm/2FIGURES/2ATP/22ComplexeMgATP.htm";
namesArray[arraycount] = "Structure et stabilisation de l'ATP";
descArray[arraycount] = "stabilization repulsion oxygene oxygen atp";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/1VELATP.h
tm/2FIGURES/2ATP/11StructHydrolATP.htm";
namesArray[arraycount] = "Structure et hydrolyse de l'ATP";
descArray[arraycount] = "hydrolyzis amp adp atp ester phosphoanhydride anhydride
phosphate inorganique inorganic";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/1VELATP.h
tm/2FIGURES/2ATP/44PhosEnolPyr.htm";

namesArray[arraycount] = "Structure et hydrolyse du PEP";
descArray[arraycount] = "phosphoenolpyruvate pyruvate pep enol phosphate";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/222OxydoRedNAD.htm";
namesArray[arraycount] = "Oxydo-reduction - Relation Nernst - NAD(P)+ - Chaine
respiratoire - Theorie chimio-osmotique - Synthese ATP";
descArray[arraycount] = "oxydation potentiel standard redox electrode hydrogene
hydrogen nad nadp nicotinamide adenine adenin dinucleotide phosphate chain
respiratory complex theory chimioosmotique chimio osmotique osmotic mitchell
gradient proton interstitiel intermenbranaire internal atp synthesis synthase atpase f0
f1 boyer walker force proton motrice animation shockwave flash";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/1ElectrodeH2/11FonctionPile.htm";
namesArray[arraycount] = "Fonctionnement electrochimique d'une pile";
descArray[arraycount] = "";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/2EqNERNST/1DemoEqNERNST.htm";
namesArray[arraycount] = "Demonstration de l'equation de NERNST";
descArray[arraycount] = "";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/3NADetNADP/11StructNadNadp.htm";
namesArray[arraycount] = "Structure nicotinamide adeninine dinucleotide
(phosphate) - NAD(P)+ forme oxydee et reduite";
descArray[arraycount] = "oxydized reduced";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA

D.htm/3FIGURES/4ReacOxRedNADP/1OxRedNad.htm";
namesArray[arraycount] = "Oxydo-reduction nicotinamide adenine dinucleotide
(phosphate) - NAD(P)+";
descArray[arraycount] = "mecanisme mechanism capture hydrure";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/4ReacOxRedNADP/4STEREOOxRedNad.htm";
namesArray[arraycount] = "Stereospecificite transfert ion hydrure reaction
oxydo-reduction NAD(P)+";
descArray[arraycount] = "stereospecificity";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/3NADetNADP/22SpecAbsorNad.htm";
namesArray[arraycount] = "Spectre d'absorbance formes oxydee et reduite
NAD(P)+";
descArray[arraycount] = "absorption spectrum";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/2MITOCHONDRIES/111StructMITO.htm";
namesArray[arraycount] = "Structure de la mitochondrie et localisation de la chaine
respiratoire";
descArray[arraycount] = "mitochondrial chain respiratory membrane";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/3ProtChRESP/2ComplexeI/3StrucFadFmn.htm";
namesArray[arraycount] = "Structure de la flavine adenine dinucleotide et de la
flavine mononucleotide";
descArray[arraycount] = "fad fmn";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA

D.htm/3FIGURES/5ChaineRespir/3ProtChRESP/4ComplexeIII/3UBIQUINONE.htm";
namesArray[arraycount] = "Les trois niveaux d'oxydation de l'ubiquinone ou
coenzyme Q";
descArray[arraycount] = "semi quinone mobile";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/4NAVETTES/1NavGLYPHOS.htm";
namesArray[arraycount] = "La navette glycerol phosphate";
descArray[arraycount] = "glycerol navette shuttle complex";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/4NAVETTES/3NavAspMal.htm";
namesArray[arraycount] = "La navette malate - aspartate";
descArray[arraycount] = "malate navette shuttle complex";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/222OxydoRedNAD.htm";
namesArray[arraycount] = "Oxydo-reduction - Relation de Nernst - NAD(P) - Chaine
respiratoire - Theorie chimio-osmotique - Synthese ATP";
descArray[arraycount] = "cytochrome ubiquinone";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/3ProtChRESP/1QuatreCOMPLEXES/11CompProt
ChRESP.htm";
namesArray[arraycount] = "Complexes proteiques de la chaine respiratoire";
descArray[arraycount] = "cytochrome complex";
categoryArray[arraycount] = "";
targetArray[arraycount] = "target=_top";
arraycount += 1;
urlsArray[arraycount] =
"http://ead.univ-angers.fr/~jaspard/Page2/COURS/1VELATPOxRedNAD/2OxRedNA
D.htm/3FIGURES/5ChaineRespir/3ProtChRESP/3ComplexeII/1MecTRANSFelec.ht
m";
 6
6
 7
7
 8
8
 9
9
 10
10
 11
11
 12
12
 13
13
 14
14
 15
15
 16
16
 17
17
 18
18
 19
19
 20
20
 21
21
 22
22
 23
23
 24
24
 25
25
 26
26
 27
27
 28
28
 29
29
 30
30
 31
31
 32
32
 33
33
 34
34
 35
35
 36
36
 37
37
 38
38
 39
39
 40
40
 41
41
 42
42
 43
43
 44
44
 45
45
 46
46
 47
47
 48
48
 49
49
 50
50
 51
51
 52
52
 53
53
 54
54
 55
55
 56
56
 57
57
 58
58
 59
59
 60
60
 61
61
 62
62
 63
63
 64
64
 65
65
 66
66
 67
67
 68
68
 69
69
 70
70
 71
71
 72
72
 73
73
 74
74
 75
75
 76
76
 77
77
 78
78
 79
79
 80
80
 81
81
 82
82
 83
83
 84
84
 85
85
 86
86
 87
87
 88
88
 89
89
 90
90
 91
91
 92
92
 93
93
 94
94
 95
95
 96
96
 97
97
 98
98
 99
99
 100
100
 101
101
 102
102
 103
103
 104
104
 105
105
 106
106
 107
107
 108
108
 109
109
 110
110
 111
111
 112
112
 113
113
 114
114
 115
115
 116
116
 117
117
 118
118
 119
119
 120
120
 121
121
 122
122
 123
123
 124
124
 125
125
 126
126
 127
127
 128
128
 129
129
 130
130
 131
131
 132
132
 133
133
1
/
133
100%